Module 3: Overview
This module’s directory (“./modules/m3_output”) contains a set of tutorials based on short set (a few weeks) of station and binary model output. These are stored in a directory named ./outputs along with supplementary files (like the global_to_local mapping files) needed to make sense out of them.
Note
There are library imports from schimpy in this module. Be sure to have installed the python schimpy package (see Getting Started - Python)
The station input file “station.in” used to launch SCHISM is how the x-,y-, and z- coordinates are associated with the station name. The z-coordinate in this example uses -1.0 m as the “default” observation depth, other depths are classified as “upper” or “lower” depending on where they are in the water column.
In the Hello SCHISM example model, there are 6 stations that have been designated prior to running the model in the “station.in” file which are distributed in the model domain:
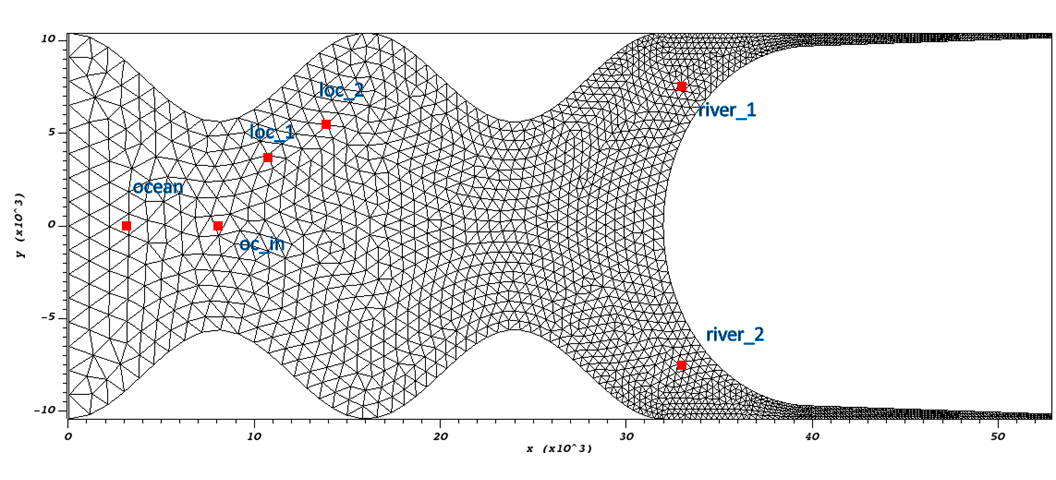
Fig. 4 Hello SCHISM observation stations as specified by the station.in file
Station Output
Exercise 1-1 : Station Elevation
In this exercise you will read and plot elevation data at a station using the SCHISM outputs provided.
Open ‘.m3_output/exercises/exercise1.py’ in a text editor (ex: Notepad++) or your Python IDE of choice (ex: Spyder, PyCharm, VSCode, etc.) Save the file as “./m3_output/exercises/exercise1-1.py”.
While making edits, ensure consistency of indentation with previous lines (there are four spaces at the front of each line below the if __name__ == ‘__main__’: block).
In this example we want to read elevation data from the San Francisco station.
Since we’re wanting the elevation data, you need to set the output folder. Add the following line at the end of the code within the if __name__ == ‘__main__’: block:
outputs_fpath = "../outputs/staout_1"
Now, to retrieve the Ocean Boundary Inland station you need to set the station_name variable as well as the sub_loc variable. These both correspond to the station.in SCHISM input file. The sub_loc corresponds to the observation depth at the station. Add the following lines after the last edit you made:
station_name = "oc_in" # Ocean Boundary Inland
sub_loc = "default"
The schimpy function read_staout is a Python data extracting function that pulls out time series from the right column of the staout_* files. Calling this function stores the requested time series for all stations. Add the following line after the last edit you made, keeping consistent with indentation:
all_ts = read_staout(outputs_fpath, station_fpath, time_basis)
To retrieve a specific station’s time series and then plot using the matplotlib plotting library in Python, add the following two lines after the last edits you made, again keeping consistent with indentation:
all_ts[station_name + "_" + sub_loc].plot()
plt.show()
Save the file (“.m3_output/exercises/exercise1-1.py”) and run the script in the command line.
python exercise1-1.py
This will open a pop-out plot of the elevation timeseries for the Ocean Inland station, you can zoom around in the plot window to evaluate the data.
Writing Out Station Data
Now if we want to write the paired timeseries data to a csv we can use pandas built-in “to_csv” function on the “all_ts[station_name + “_” + sub_loc]” object (which is a pandas Series object).
Open your saved exercise1-1.py script.
Comment out the all_ts[station_name + “_” + sub_loc].plot() and plt.show() lines by adding a “#” before each line.
Add a line to write the series out to a csv using the following command:
all_ts[station_name + "_" + sub_loc].index.rename('Datetime', inplace=True)
all_ts[station_name + "_" + sub_loc].to_csv("oc_in_elevation_ts.csv", index=True, header=['Elevation (m)'])
This will rename the index of the series to “Datetime” so that the csv header uses “Datetime” for the index column, and then it writes out the csv file oc_in_elevation_ts.csv while specifying that the data column has the header of “Elevation (m)”.
Take a look at the csv file created and see that it’s written out data that looks like this:
Datetime,Elevation (m)
2009-02-10 00:06:00,1.0
2009-02-10 00:12:00,1.00022
2009-02-10 00:18:00,1.00176
Exercise 1-2 : Station Salinity
In this exercise you will read and plot salinity data at a different station than Exercise 1-1.
Re-Open ‘./m3_output/exercises/exercise1.py’. This should be the raw/original script without any edits you made in Exercise 1-1. Save this file as “./m3_output/exercises/exercise1-2.py”.
In this exercise we want to evaluate salinity data from the station at Location 1 (Fig. 4).
Since we’re pulling salinity data, you need to specify the output folder. Add the following line at the end of the code within the if __name__ == ‘__main__’: block:
outputs_fpath = "../outputs/staout_6"
Now, to retrieve the River 1 station you need to set the station_name variable as well as the sub_loc variable. Add the following lines after the last edit you made:
station_name = "loc_1" # Location 1
sub_loc = "upper"
The schimpy function read_staout is a Python data extracting function that pulls out time series from the right column of the staout_* files. Calling this function stores the requested time series for all stations. Add the following line after the last edit you made, keeping consistent with indentation:
all_ts = read_staout(outputs_fpath, station_fpath, time_basis)
To retrieve a specific station’s time series and then plot using the matplotlib plotting library in Python, add the following two lines after the last edits you made, again keeping consistent with indentation:
all_ts[station_name + "_" + sub_loc].plot()
plt.show()
Save the file (“./m3_output/exercises/exercise1-2.py”) and run the script in the command line.
python exercise1-2.py
This will open a pop-out plot of the salinity timeseries for Location 1, you can zoom around in the plot window to evaluate the data.
(Optional) Retrieve salt at the same station with ‘sub_loc=lower’ and plot it with the previous time series. You can do this by adding the following line before you call plt.show()
all_ts[station_name + "_lower"].plot()
Exercise 2 : Transect Flow
In this exercise you will read and plot flow data using the flux.out file found in the ./outputs folder as well as the flowlines.yaml file found in the folder for this exercise “./m3_output”.
Open ‘./m3_output/exercises/exercise2.py’
Since we want to retrieve flow data, you need to specify the output path as well as the . Add the following lines at the end of the code within the if __name__ == ‘__main__’: block:
station_fpath = '../flowlines.yaml'
outputs_fpath = "../outputs/flux.out"
For this exercise you will read data from the mouth of River 1, so set the station_name using the following line:
station_name = "north_weir_upstream"
The schimpy function read_flux_out is a Python data extracting function that pulls out time series from the right column of the flux.out file. Calling this function stores the requested time series for all observation flow stations. Add the following line after the last edit you made, keeping consistent with indentation:
all_ts = read_flux_out(outputs_fpath, station_fpath, time_basis)
To retrieve a specific station’s time series and then plot using the matplotlib plotting library in Python, add the following two lines after the last edits you made, again keeping consistent with indentation:
all_ts[station_name].plot()
plt.show()
Save the file (“./m3_output/exercises/exercise2.py”) and run the script in the command line.
python exercise2.py
This will open a pop-out plot of the flow timeseries from just upstream of River 1’s weir, you can zoom around in the plot window to evaluate the data.
Exercise 3 : Observed Data
In this exercise you will read and plot an observation file not created by SCHISM, but a paired timeseries found in “./m3_output/exercises/9414290_gageheight.txt”. If you open the file in a text editor (ex: Notepad++) you can see that there is a Date Time column, a Water Level column and some other quality flags. This timeseries was obtained from NOAA and has 7 lines of commented-out material, one header line and then the timeseries data.
Open ‘./m3_output/exercises/exercise3.py’ in a text editor or Python IDE.
Specify the input file by entering the following below the last line, keeping the indentation consistent with the above lines.
fpath_in = '9414290_gageheight.txt'
Gather the data into a “ts” object by entering the following line next:
ts = read_ts(fpath_in)
Plot the data by adding these two lines at the end of the script:
ts.plot()
plt.show()
Save the file (“./m3_output/exercises/exercise3.py”) and run the script in the command line.
python exercise3.py
This will open a pop-out plot of the elevation timeseries, you can zoom around in the plot window to evaluate the data.
Exercise 4 : Combined Plots
In this exercise you will combine techniques found in Exercise 1-1 and Exercise 3 to compare two time series in a plot.
Open ‘./m3_output/exercises/exercise4.py’
Use the techniques in Exercise 1-1 to define the outputs_fpath, station_name, and sub_loc variables so that you get the elevation data you retrieved previously. Write the function to store this data into a variable called “sim_data”.
Now, enter an empty line to differentiate the two data retrieval inputs. Use the techniques in Exercise 3 to gather the same observation data into a variable called “ts_obs”. Recall, you’ll need to define the obs_fpath variable.
The ts_obs object now has the ability to rename the column so that when plotting in matplotlib the legend will reflect an aptly named column. Use the following line to change the name of the “Water Level” column to “Obs”:
ts_obs = ts_obs.rename(columns={"Water Level":"Obs"})
Now, to plot the time series with legends, use the following lines of code after entering an empty line to imply a new block of plotting commands.
ts_obs.plot(legend=True, ax=ax)
sim_data[station_name+"_"+sub_loc].plot(label="Sim", legend=True, ax=ax, linestyle='dashed')
plt.show()
Save the file (“./m3_output/exercises/exercise4.py”) and run the script in the command line.
python exercise4.py
This will open a pop-out plot of the elevation timeseries, you can zoom around in the plot window to evaluate the data
Binary Output
Previously, you used the station output files and observed timeseries files to gather and plot data. Now you will use binary outputs to extract and plot timeseries. This is what you’d typically need to do if you hadn’t set up the output station and transect locations prior to running the model.
Navigate to folder output, and create ‘station.bp,’ which has extraction points. Let’s add one station.
mid.bp
1 ! # of stations to extract
1 20000. 0. -1.0 ! Middle
The first line is just comment, the second line is the number of stations, and the third is the list of station x-, y-, and z-coordinates. Notice the z-coordinate -1.0 in the third line, the post-processsing script is capable of interpreting this value as either an absolute elevation relative to the model datum (ex: -1.0 m NAVD88) or a relative elevation to the surface.
Warning
This explanation needs to be updated.
We will treat it as absolute elevation later by specifying vertical coordinate system by screen inputting. ((((Will we?)))) Most of water column in our Bay-Delta model are negative. You can also use depth from free surface instead as vertical location, which is defined positive from surface to bottom.
Run ‘read_output10_xyz’ in the command line, and input parameter after each bold screen inquiry.
read_output10_xyz
Input extraction pts format (1: .bp; 2:.sta):
1
Input ics (1-linear interp; 2-nearest neighbor interp. 2 for node-based variables only! 2 is suggested for sub-meter resolution!):
1
Input variable name to read from nc (e.g. elevation):
elevation
Is the var node (1) or elem (2) based?
1
Input start and end file # to read:
22 25
Is the z-coord. in station.* relative to surface (1) or a fixed level (0)?
0
The result will be saved in ‘fort.18’ in this case, take a look at it with Excel and plot the time series.
Warning
This doesn’t actually work for me. It doesn’t seem to be able to find the station.bp file even when in the right directory.
(Optional) Add more stations and repeat the exercise.